The modular architecture of P2TF is described in the following figure.
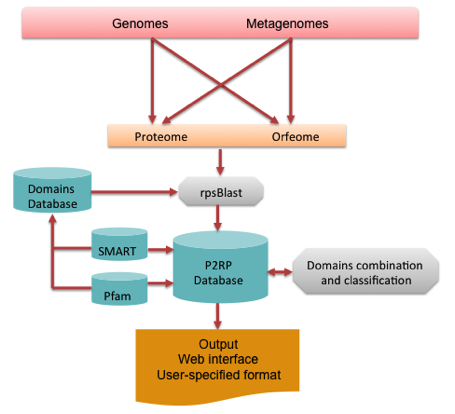
Prokaryotic TFs were defined through a comprehensive of computational analysis completely sequenced genomes and metagenomes. The P2TF pipeline is designed to take protein and whole replicon genomic files as input. In the second case the DNA sequence is scanned to predict the entire set of valid ORFs, defined as the DNA segments occurring between two stop codons and exceeding 100 nucleotides, with no further assumption about the presence or not of coding sequences. Thereafter the sequences are translated to constitute the ORFeome. This approach allows identification of possibly mis-predicted (overlooked) TF proteins, by comparison to the predicted TF from the proteome pool.
P2TF predicts TF candidates by performing domain analysis of each protein sequence. We manually selected a pool of domains from the Pfam and SMART libraries, based on analysis of the literature on sequence-specific DBDs and their associated domains. The presence in a protein of a domain defined as a DBD led to classification of the protein as a TF and inclusion in P2TF.
The P2TF analysis then divides DBDs into TFs and ŌOther DNA-binding ProteinsÕ (ODPs), a category which includes non-regulatory DNA-binding proteins such as tranposases, integrases and histone-like proteins. TFs are then further divided into sub-categories (Transcriptional Regulators (TRs) including OCSs, RRs and SFs) according to their domain architecture. TFs which contain a CheY-like receiver (phosphoacceptor) domain and annotates them as RRs. These proteins form, with partner sensor histidine kinases, two-component systems, which are specifically analysed elsewhere. SFs were divided into 3 sub-groupings; RpoN, RpoD and ECF (extra-cytoplasmic function) SFs. OCSs were defined as proteins carrying both input and output domains but lacking phosphotransfer domains characteristic of two-component systems.
TFs, OCSs and RRs were also divided into 66 distinct sub-groupings (families) depending upon which domains were present in the proteins.